Model data¶
metawards is a geographical SIR model. This means that a separate
progression of states from S through to R is performed in every
ward that is modelled. A ward is just a geographic area or cell.
It originally represented a single electoral ward in the UK (hence the name),
but there is no technical reason why this has to be the case.
Infected individuals who are resident in a ward contribute to the force of infection (FOI) of that ward. The higher the FOI, the more likely it is that another resident will be infected.
In addition, some residents (called workers) make regular movements each day between their home ward and a work ward. They contribute to and are exposed to the FOI of their work ward, and so can spread the infection between wards.
The number of wards, geographic data, populations and population flows
between them are altogether called the model data. metawards was
first developed to model electoral wards in England and Wales, with
data based on the 2011 census. The default model used in metawards
is thus this 2011Data model.
You can set the model used by metawards using the --model or -m
command line argument, e.g.
metawards -d lurgy3 -m 2011Data
would perform a model run using your new lurgy3 parameters on the default
2011Data model data. We have been, and will continue to use
this 2011Data default model for
all of the examples in this tutorial. However, please remember that
you can create your own model data to represent any location, region,
country or interacting sets of groups as you wish.
Alternative models¶
Constructing this model data involves a lot of data science and careful consideration. The models are held in the MetaWardsData repository, and the current models include;
2011Data: default data for England and Wales from the 2011 census2011UK: work-in-progress model for the whole of the UK including Northern Ireland.
In addition, there is a special model called single, that is used to
perform validation model runs where the entire population is resident
in only a single ward. This is useful if you want to validate the underlying
models used in metawards, or want to see if results obtained are
influenced by geography.
You should specify the size of the population when you use the single
model via the --population or -P keyword, e.g.
metawards -d lurgy3 -a ExtraSeedsOne.dat -m single -P 50000000 --nsteps 2000
would perform a run with 50 million individuals in a single ward.
Note
We have used ExtraSeedsOne.dat to seed this run, as this file
seeds 5 infections into the first ward of a model on day 1. We’ve
also set nsteps to 2000 to stop the model timing out after
two years. This is because the lack of geographic mixing significantly
slows the progression of the disease.
After running this command, you can create an overview plot using;
metawards-plot -i output/results.csv.bz2
The output from your run should look something like this;
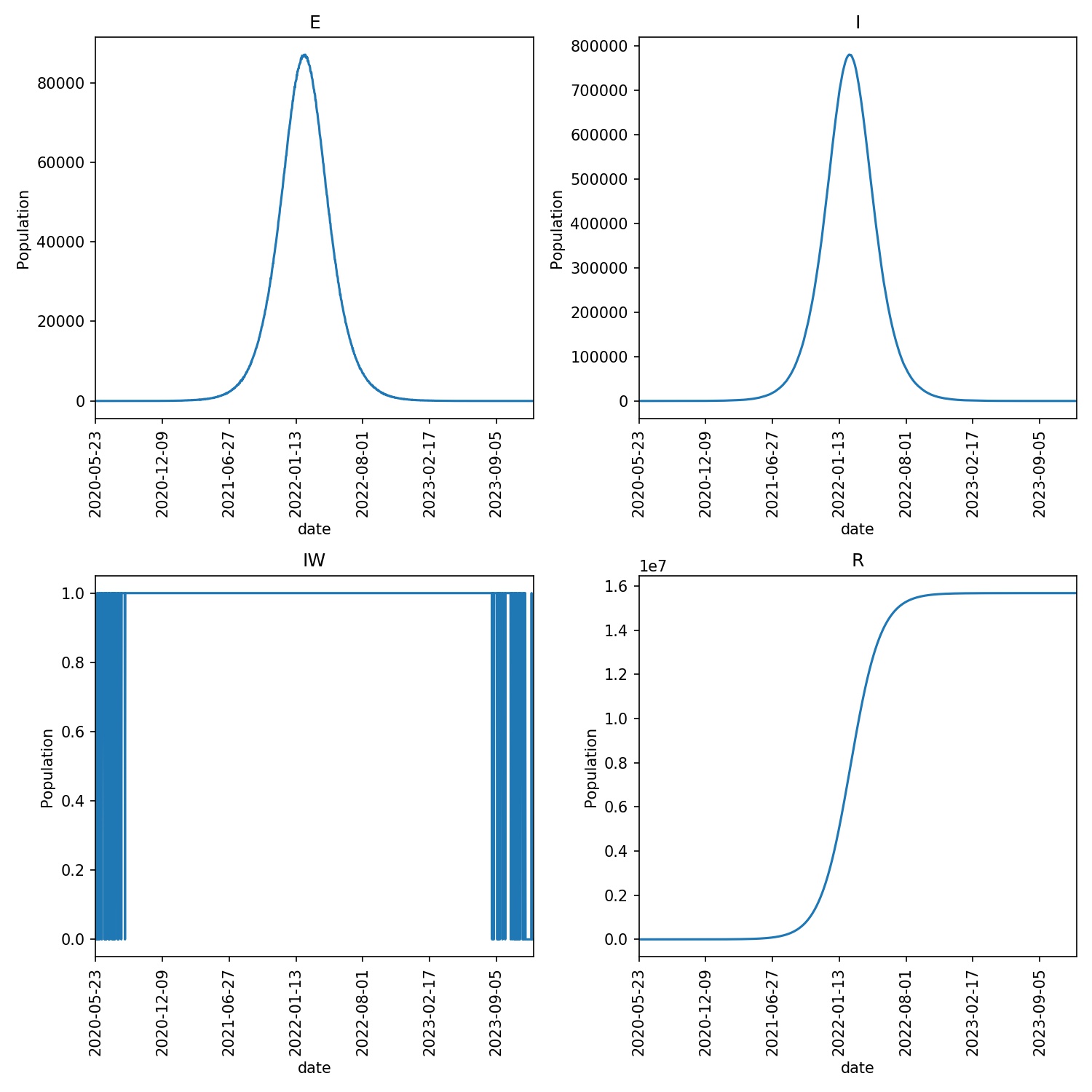
Note
You may find that the disease dies out quickly in a small number of runs. This is due to the random nature of the model, and random chance means that the disease in those runs could not catch light. If it does catch, then the epidemic should progress for ~1000 days.