How many days of self-isolation is needed?
In the last page you saw that about 1000 individuals were still
infected after seven days of self-isolation, and were released
back into the community. This was just 1-1.5% of the total number of
infections, so it was unlikely to have a big impact. We can
investigate this impact by scanning the number of days of
self-isolation required. Edit your move_isolate.py to read;
from metawards import Population
from metawards import Networks
from metawards.movers import go_isolate, go_to
from metawards.utils import Console
def move_isolate(network: Networks, population: Population, **kwargs):
user_params = network.params.user_params
ndays = user_params["isolate_ndays"]
isolate_stage = user_params["isolate_stage"]
if ndays > 7:
Console.error(f"move_isolate supports a maximum of 7 days of "
f"isolation, so {ndays} is too many!")
raise ValueError("Too many days of isolation requested")
elif ndays <= 0:
# just send infected individuals straight to "released"
# (this is the control)
func = lambda **kwargs: go_isolate(
go_from="home",
go_to="released",
self_isolate_stage=isolate_stage,
**kwargs)
return [func]
day = population.day % ndays
isolate = f"isolate_{day}"
go_isolate_day = lambda **kwargs: go_isolate(
go_from="home",
go_to=isolate,
self_isolate_stage=isolate_stage,
**kwargs)
go_released = lambda **kwargs: go_to(go_from=isolate,
go_to="released",
**kwargs)
return [go_released, go_isolate_day]
This move function will now read the number of days to isolate from
the isolate_ndays user parameter. It will also move an individual
into self-isolation when they reach disease stage isolate_stage.
There is a little error-catching, e.g. as we only have seven available
isolate_N demographics, we can only model up to seven days of
self-isolation. If more than seven days is requested this calls
Console.error to write an
error to the output, and raises a Python ValueError.
To add a control, we’ve set that if isolate_ndays is zero, then
infected individuals are sent straight from home to released.
This way we can compare runs that use self-isolation against a run
where self-isolation is not performed.
Note
We could of course increase the number of days of self-isolation that
could be modelled by editing demographics.json and
mix_isolate.py and just increasing the number of isolate_N
demographics.
Next create a file called scan_isolate.dat and copy in;
# Scan through self-isolation from 0 to 7 days,
# with self-isolation starting from disease
# stages 2-4
.isolate_stage .isolate_ndays
2 0
2 1
2 2
2 3
2 4
2 5
2 6
2 7
3 0
3 1
3 2
3 3
3 4
3 5
3 6
3 7
4 0
4 1
4 2
4 3
4 4
4 5
4 6
4 7
This file will scan isolate_stage from 2 to 4, while scanning
isolate_ndays from 0 to 7.
You can run this job locally using the command;
metawards -d lurgy4 -D demographics.json -a ExtraSeedsLondon.dat --mixer mix_isolate --mover move_isolate --extractor extract_none --nsteps 365 -i scan_isolate.dat
Given that it is good to repeat the runs several times, and there are a lot of jobs, you may want to run this on a cluster. To do this, the PBS job script could look like;
#!/bin/bash
#PBS -l walltime=12:00:00
#PBS -l select=4:ncpus=64:mem=64GB
# The above sets 4 nodes with 64 cores each
source $HOME/envs/metawards/bin/activate
# change into the directory from which this job was submitted
cd $PBS_O_WORKDIR
metawards -d lurgy4 -D demographics.json -a ExtraSeedsLondon.dat \
--mixer mix_isolate --mover move_isolate --extractor extract_none \
--nsteps 365 -i scan_isolate.dat --repeats 8 \
--nthreads 16 --force-overwrite-output --no-spinner --theme simple
while the slurm job script would be;
#!/bin/bash
#SBATCH --time=01:00:00
#SBATCH --ntasks=4
#SBATCH --cpus-per-task=64
# The above sets 4 nodes with 64 cores each
source $HOME/envs/metawards/bin/activate
metawards -d lurgy4 -D demographics.json -a ExtraSeedsLondon.dat \
--mixer mix_isolate --mover move_isolate --extractor extract_none \
--nsteps 365 -i scan_isolate.dat --repeats 8 \
--nthreads 16 --force-overwrite-output --no-spinner --theme simple
Note
Notice that we’ve set the output theme to simple using
--theme simple and have switched off the progress spinner
using --no-spinner as these are unnecessary when run in batch
mode on a cluster. All of the output will be written to the
output/console.log.bz2 file.
Analysing the results
Once the job has completed you can generate and animate the overview plots
using metawards-plot, e.g. via
metawards-plot -i output/results.csv.bz2
metawards-plot --animate -i output/overview_*.jpg
The resulting animation should look something like this;
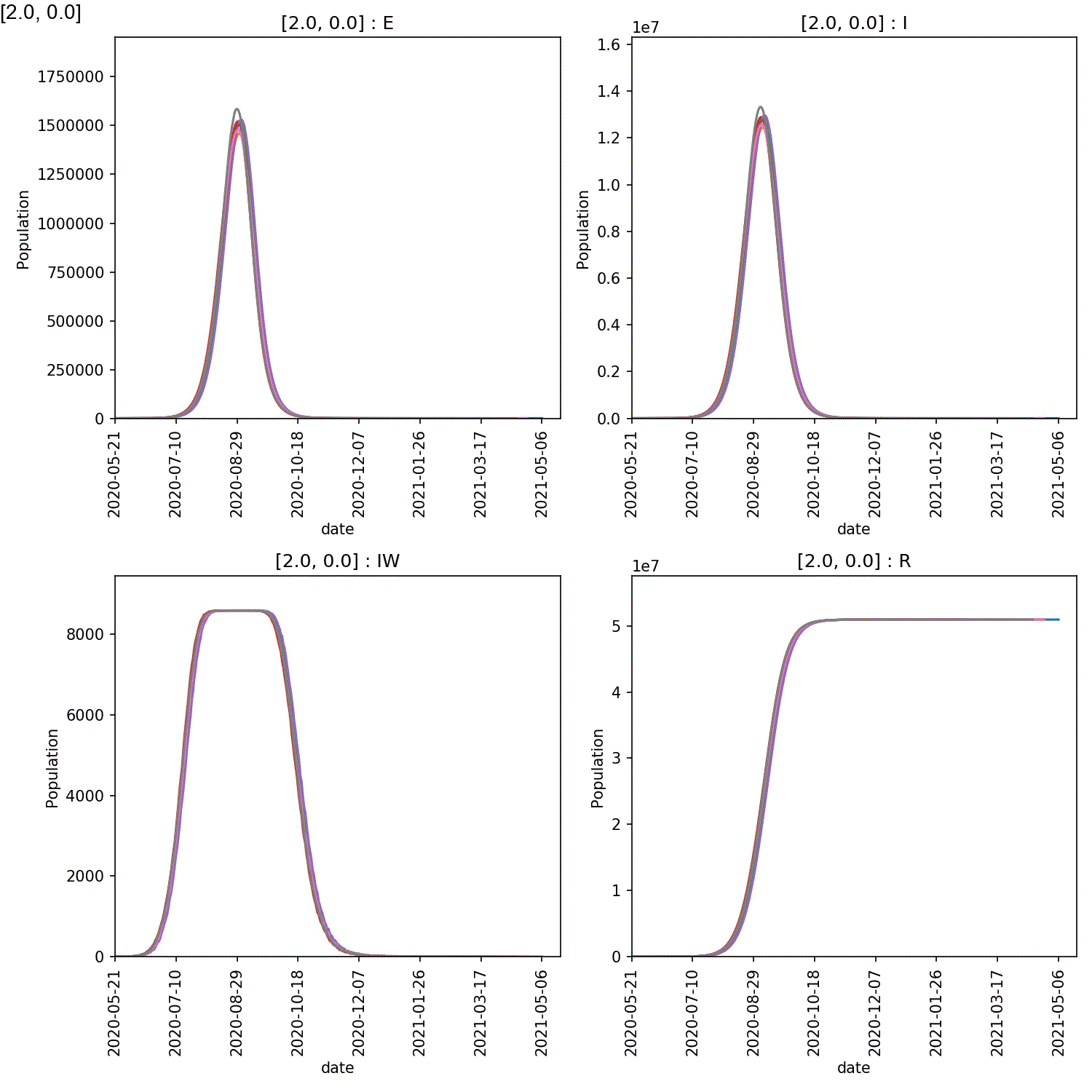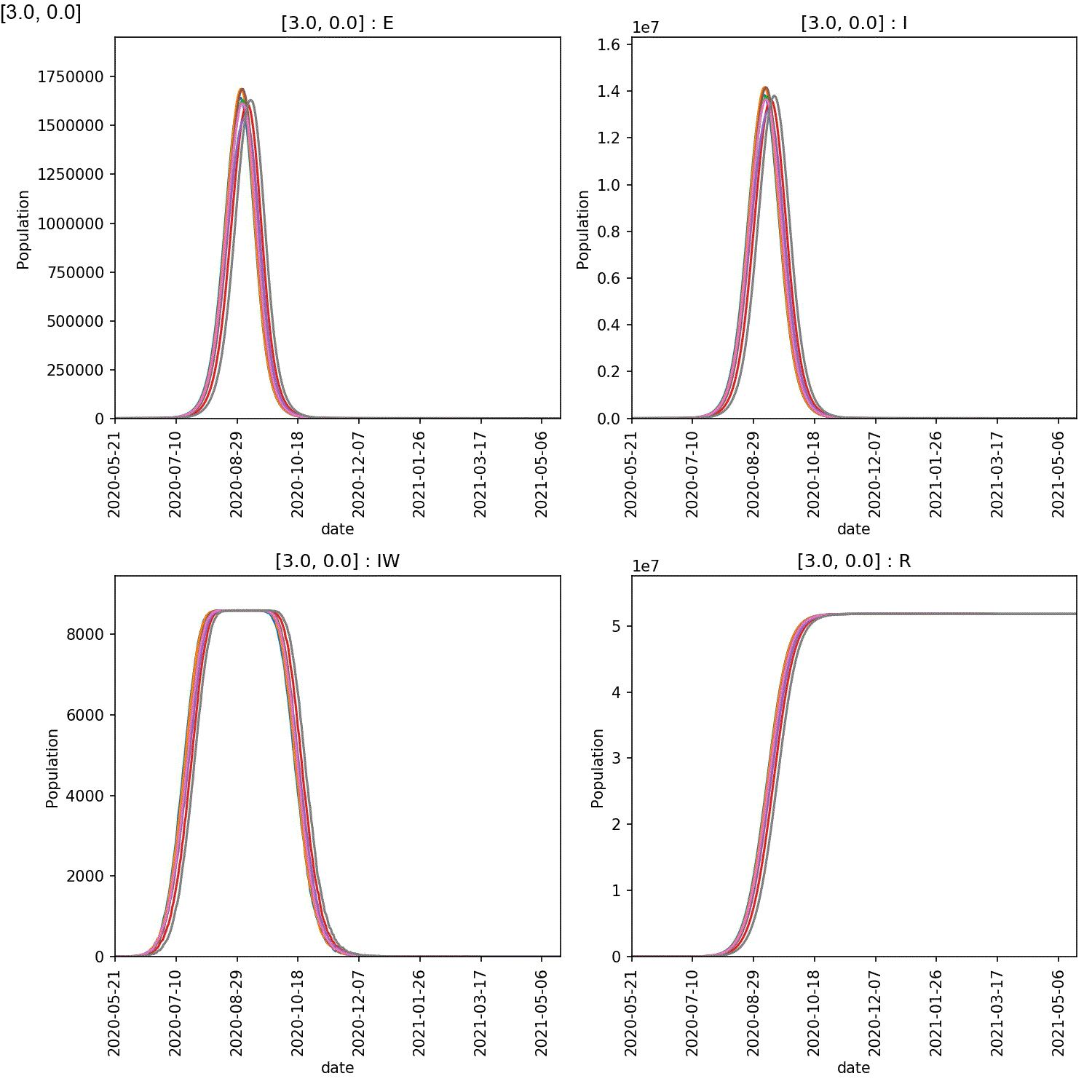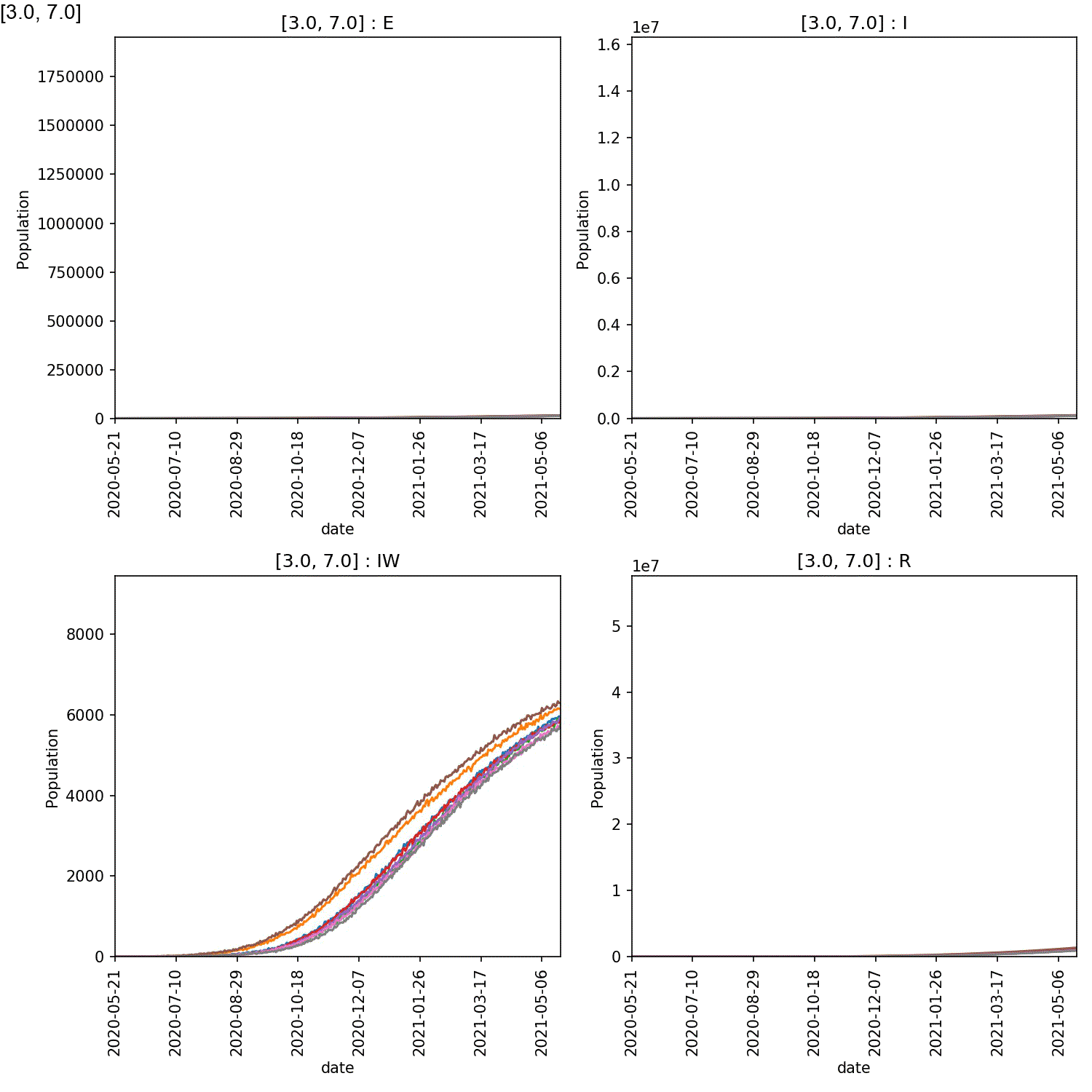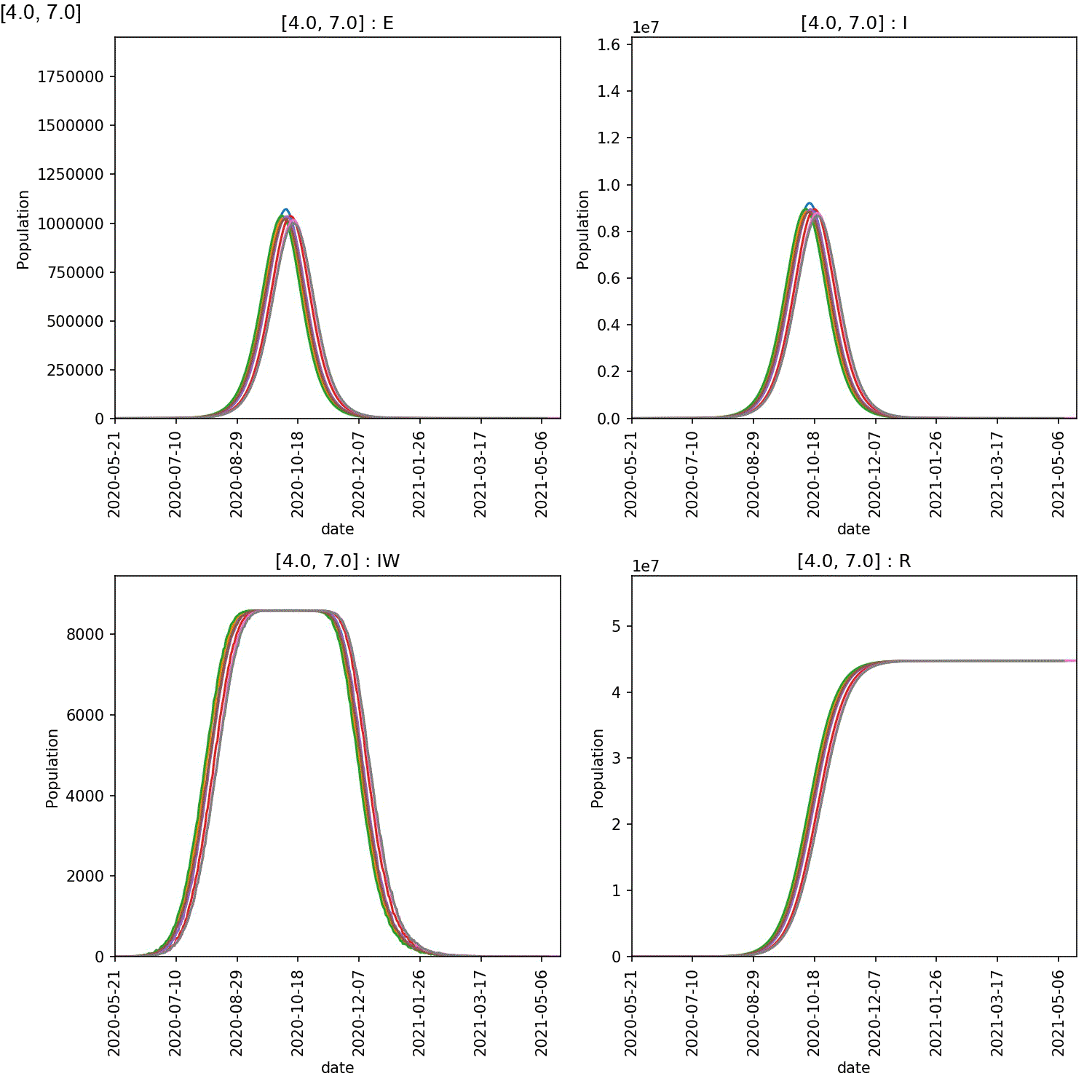
It is clear from these plots that self-isolation can work if it is started at an early stage, e.g. stage 2 or 3. Self-isolating for 5-7 days significantly reduces the numbers infected by the lurgy after a year, if started as soon as possible.
However, what is also clear is that self-isolation has little impact if it starts too late, e.g. by stage 4 of the lurgy. In this case, even using the full seven days of self-isolation, more than 40 M individuals are infected with six months.
The reason is clear when looking at the demographic plots. These can be generated using;
metawards-plot -i output/*/trajectory.csv.bz2
metawards-plot --animate -i output/*x001/demographics.jpg -o demographics.gif
Mine are shown here;
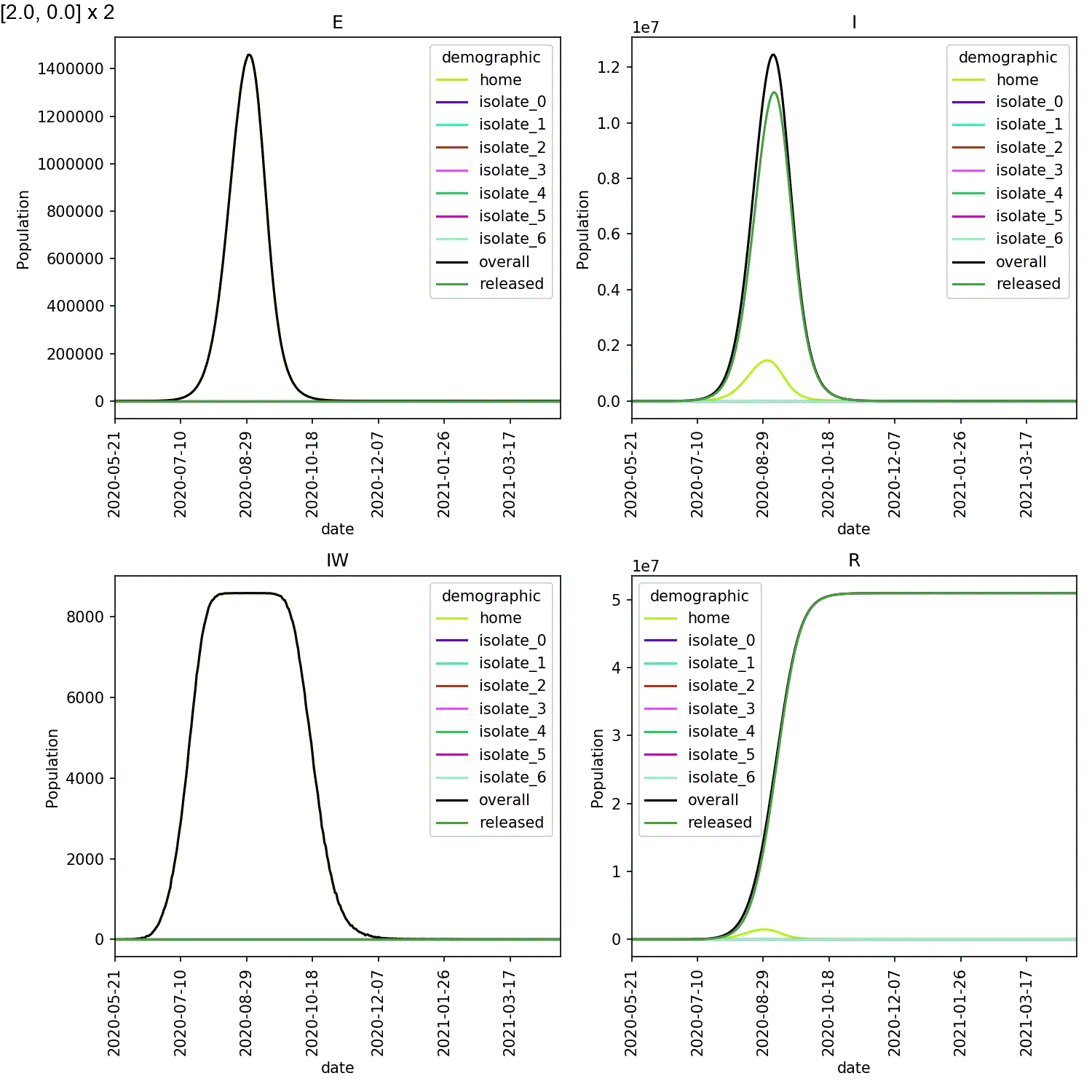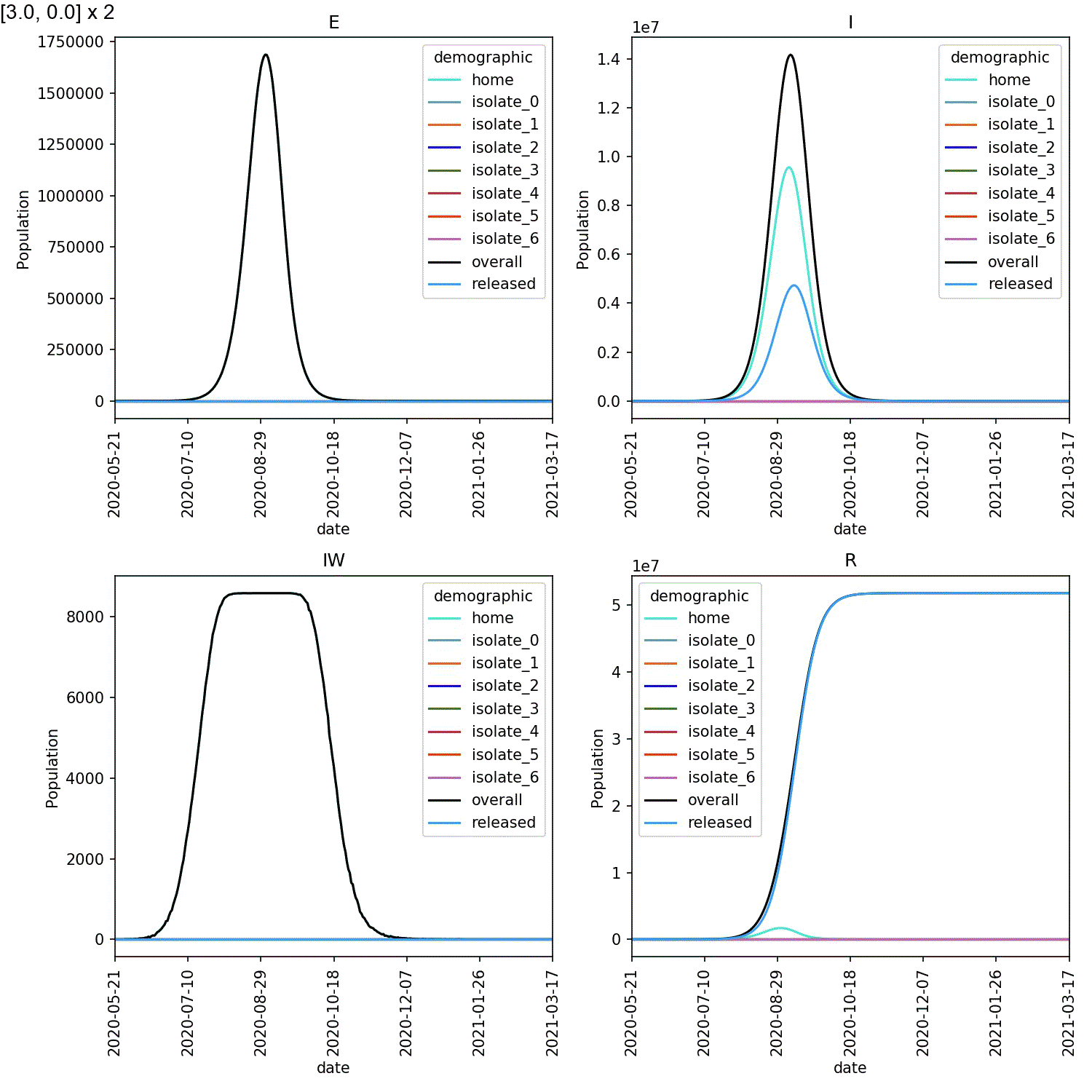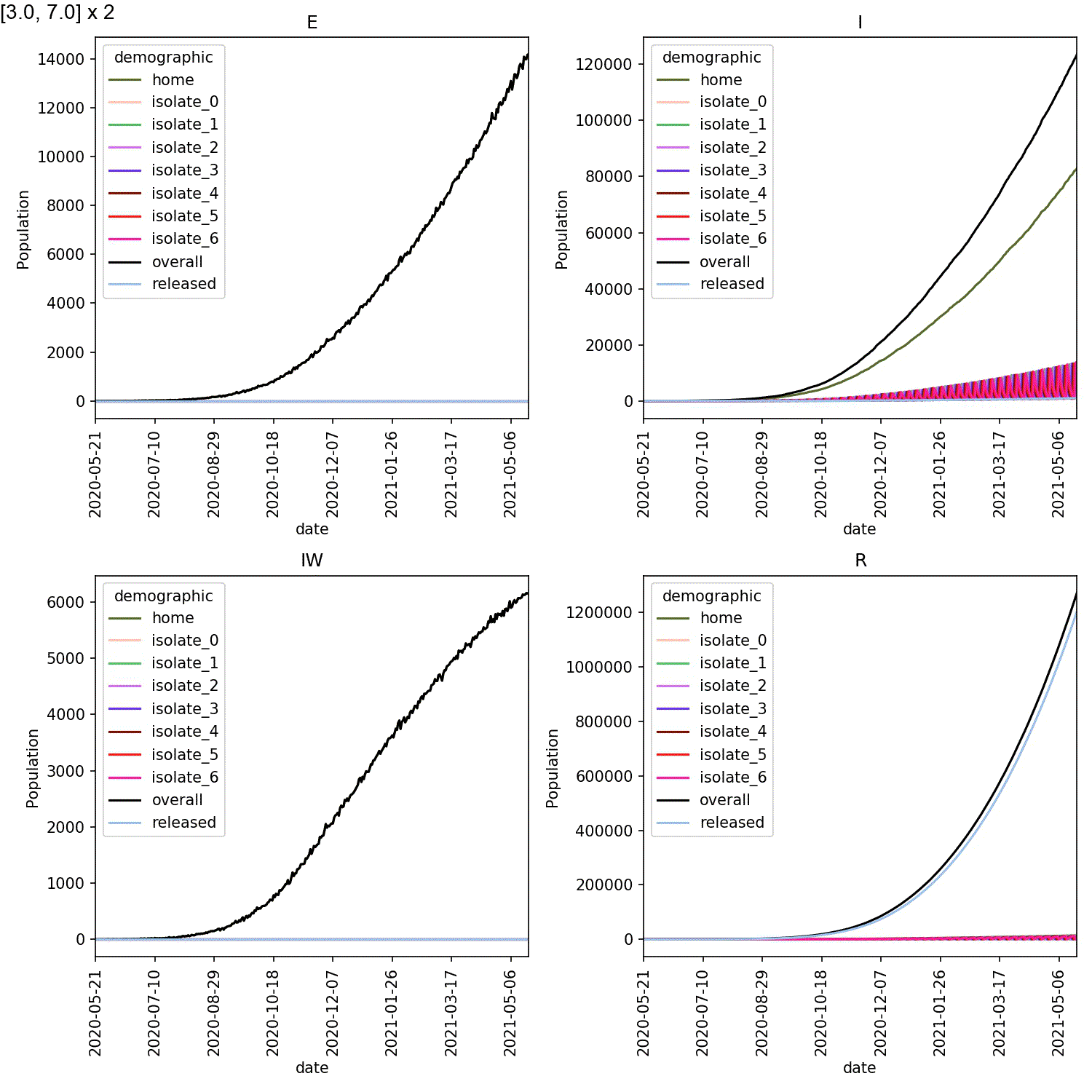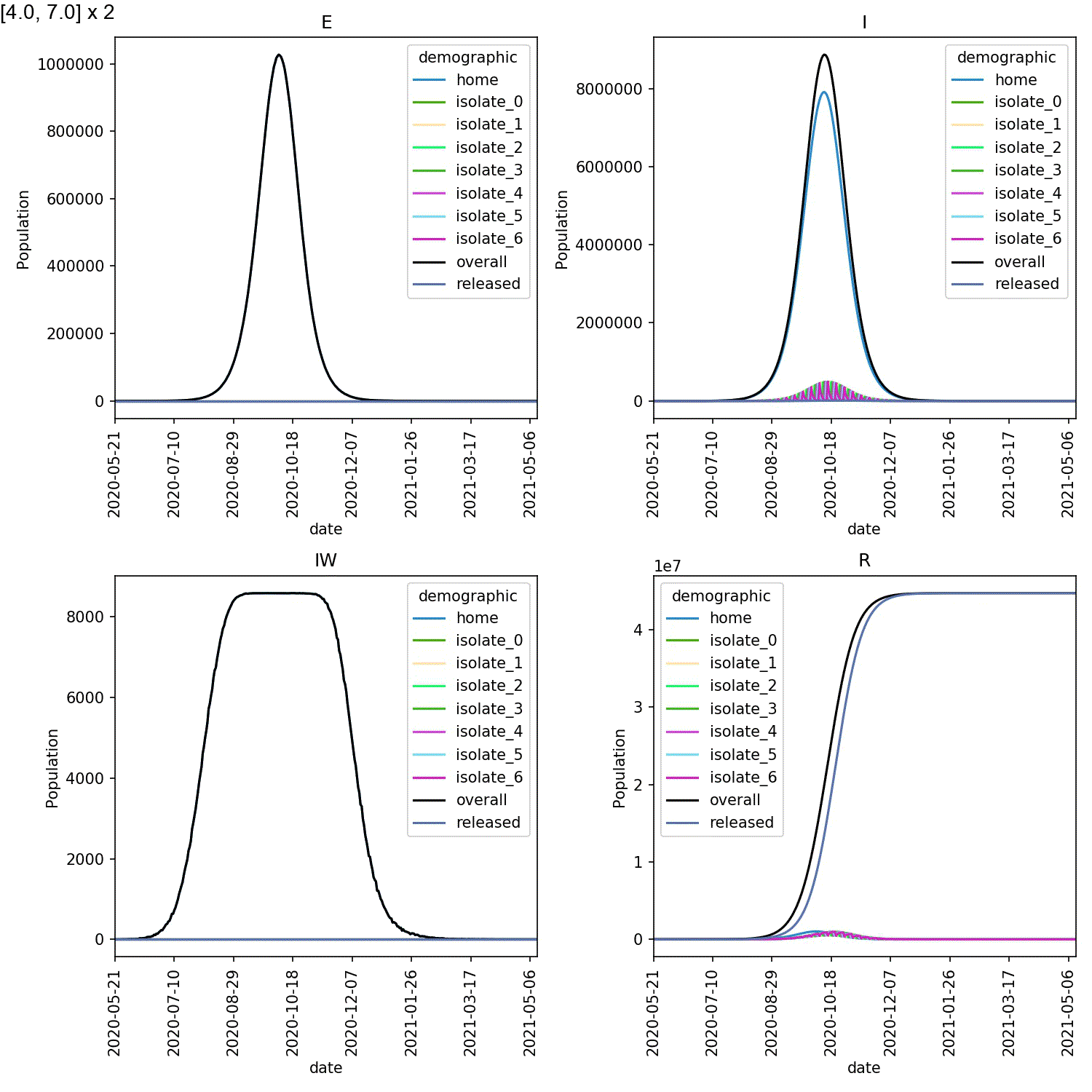
When caught early, most of the infected individuals are in one of the
isolate_N demographics. However, when starting isolation from stage 4,
the vast majority of infected individuals are in the home demographic.
The epidemic experienced exponential growth within the home demographic
before individuals self-isolated, meaning the impact of self-isolation
on the course of the outbreak was minimal.
Relections on results
For the lurgy, these results indicate that there is a very short window of time between an individual showing symptoms and electively going into self-isolation, for which self-isolation is effective.
Self-isolation (and by extensions, testing, contact-tracing etc.) would not work for the lurgy in this model if individuals waited until they’d reached stage 4 (showing larger symptoms and being more infectious), or if the time it took to confirm an infection or trace infected individuals was longer than the time it took to progress from stage 3 to stage 4.
It has to be done immediately from stage 3, when individuals start to notice symptoms (even though, at this stage, the symptoms are not limiting their movement).
It should also be noted that, in this model, beta for the asymptomatic
infectious stage had already been much reduced to account for society
adopting some control measures.