Custom advance functions
So far we have just been creating our own custom iterator functions. You can also create your own custom advance functions.
Create a new python file called advance.py and copy in the
below text;
from metawards.utils import Console
def advance_function(**kwargs):
Console.debug("Hello advance_function")
def iterate_advance(**kwargs):
Console.debug("Hello iterate_advance")
return [advance_function]
This defines two functions. One is the advance function we will use.
This takes **kwargs as arguments, but returns nothing.
The iterate_advance function is our iterator, which returns
a list of just our advance_function to run.
Use this iterator via the metawards command;
metawards -d lurgy3 --additional ExtraSeedsLondon.dat --iterator advance --debug
You should see that your “Hellos” from these functions are printed.
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ Day 0 ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━
[15:35:18] Hello iterate_advance advance.py:9
Hello advance_function advance.py:5
S: 56082077 E: 0 I: 0 R: 0 IW: 0 POPULATION: 56082077
Number of infections: 0
Creating advance_lockdown
We can write a custom advance function that represents a lockdown. To do this we will assume that there are no new work infections during a lockdown. We want to create an iterator that advances normally before the lockdown is triggered, and then advances using only the play function afterwards.
To do this, create a new python file called lockdown.py, and copy in;
from metawards.iterators import iterate_working_week, \
advance_infprob, \
advance_fixed, \
advance_play
from metawards.utils import Console
def advance_lockdown(**kwargs):
Console.debug("We are on lockdown")
advance_infprob(**kwargs)
advance_play(**kwargs)
def iterate_lockdown(population, **kwargs):
if population.day > 20:
return [advance_lockdown]
else:
return iterate_working_week(population=population,
**kwargs)
Here, the iterate_lockdown function behaves identically
to iterate_working_week for the first 20 days of the
outbreak. However, after day 20, advance_lockdown is called.
This function calls advance_infprob()
to advance the infection probabilities before calling
advance_play() to advance the
“play” infections only. This is the equivalent of making
every day into a weekend, e.g. there is no fixed or predictable
travelling from home to work (or school).
Run this iterator and draw a graph using;
metawards -d lurgy3 --additional ExtraSeedsLondon.dat --iterator lockdown
metawards-plot -i output/results.csv.bz2 --format jpg --dpi 150
You should see that the overview plot is very similar to your “weekend” only graphs that you created on the last page. This is unsurprising, as this “lockdown” has not reduced the random infections.
Scaling the infection rate
To model a reduction in the rate of new infections, the
advance_infprob() function can take
and extra scale_rate argument that is used to scale all of the
infection rates that are calculated. This is a blunt tool, but it can
be used to model the reduced infection rates that a lockdown aim to
achieve.
To set scale_rate, edit your lockdown.py file to contain;
from metawards.iterators import iterate_working_week, \
advance_infprob, \
advance_fixed, \
advance_play
from metawards.utils import Console
def advance_lockdown(**kwargs):
Console.debug("We are on lockdown")
advance_infprob(scale_rate=0.25, **kwargs)
advance_play(**kwargs)
def iterate_lockdown(population, **kwargs):
if population.day > 20:
return [advance_lockdown]
else:
return iterate_working_week(population=population,
**kwargs)
All we have done is set scale_rate=0.25 in
advance_infprob(). This represents
a four-fold reduction in the infection rate.
Run the model and generate graphs again using;
metawards -d lurgy3 --additional ExtraSeedsLondon.dat --iterator lockdown
metawards-plot -i output/results.csv.bz2 --format jpg --dpi 150
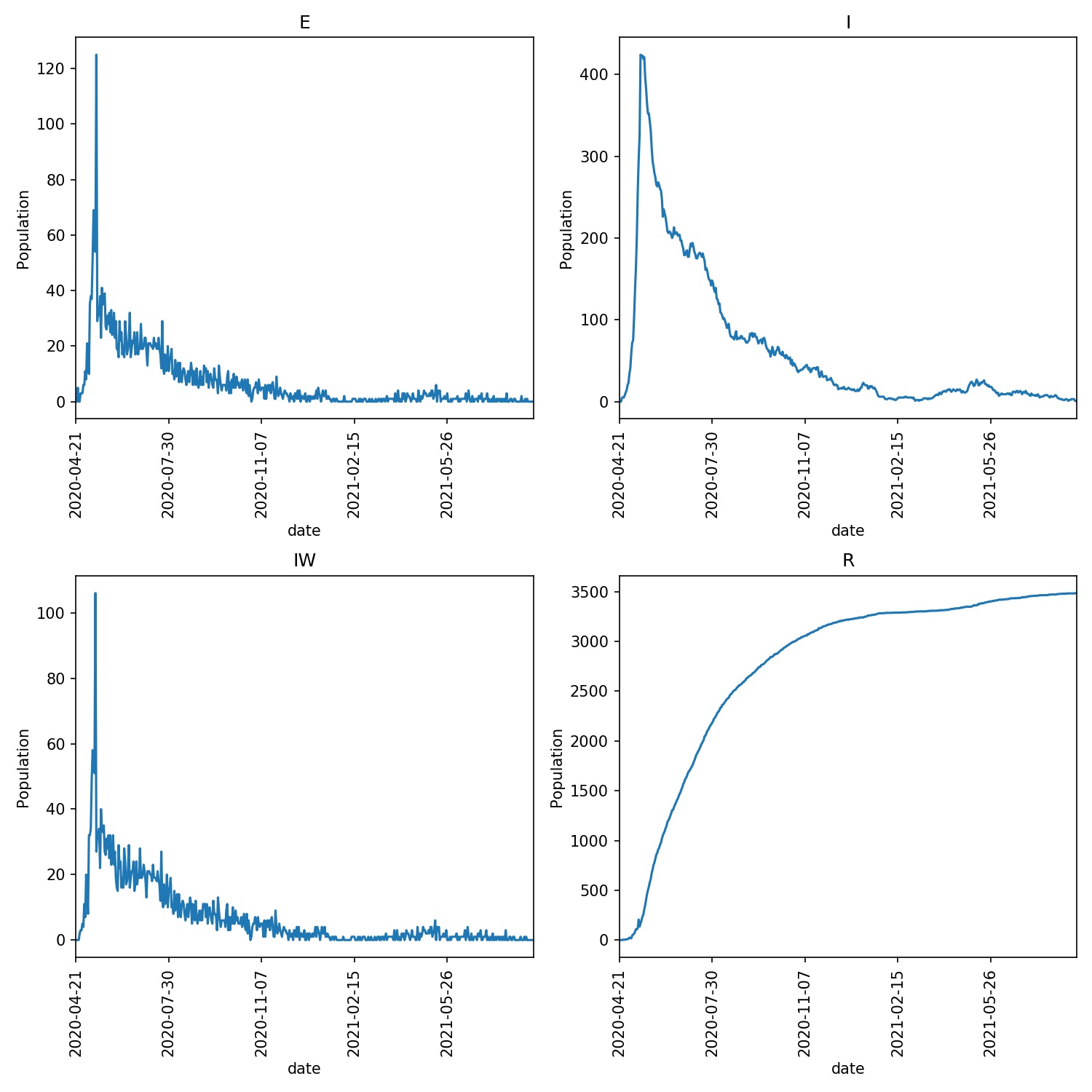
You should now see a dramatic reduction in the infection, e.g. my overview graph looks like this;