Fading immunity
Immunity to a disease is not always permanent. For some diseases, immunity fades over time, meaning that an individual can be infected multiple times.
We can model this by using go_ward() to move
individuals from the R stage back to the S stage after a period of time.
Slowing progress
The Disease.progress parameter controls
the rate at which an individual will move through a disease stage.
Advancement through the disease stages is controlled by
advance_recovery(). This simply samples
the fraction, Disease.progress,
from the number of individuals who are at that disease stage via
a random binomial distribution, and advances them to the next stage.
The key code that does this is summarised here;
# loop from the penultimate disease stage back to the first stage
for i in range(N_INF_CLASSES-2, -1, -1):
...
# get the progress parameter for this disease stage
disease_progress = params.disease_params.progress[i]
# loop over all ward-links for this disease stage
for j in range(1, nlinks_plus_one):
# get the number of workers in this link at this stage
inf_ij = infections_i[j]
if inf_ij > 0:
# sample l workers from this stage based on disease_progress
l = _ran_binomial(rng, disease_progress, inf_ij)
if l > 0:
# move l workers from this stage to the next stage
infections_i_plus_one[j] += l
infections_i[j] -= l
# loop over all nodes / wards for this disease stage
for j in range(1, nnodes_plus_one):
# get the number of players in this ward at this stage
inf_ij = play_infections_i[j]
if inf_ij > 0:
# sample l players from this stage based on disease_progress
l = _ran_binomial(rng, disease_progress, inf_ij)
if l > 0:
# move l players from this stage to the next stage
play_infections_i_plus_one[j] += l
play_infections_i[j] -= l
Time since recovery
To measure time since recovery, we can add extra “post-recovery” stages. Individuals will be set to move slowly through those “post-recovery” stages, until, when a particular post-recovery stage is reached, a fraction of individuals are deemed to have lost immunity to the disease, and are moved back to the S stage.
To do this, we will create a new version of the lurgy with these extra
post-recovery stages, which we will call R1 to R10. To do this
in Python, open ipython or jupyter and type;
>>> from metawards import Disease
>>> lurgy = Disease("lurgy6")
>>> lurgy.add("E", beta=0.0, progress=1.0)
>>> lurgy.add("I1", beta=0.4, progress=0.2)
>>> lurgy.add("I2", beta=0.5, progress=0.5, too_ill_to_move=0.5)
>>> lurgy.add("I3", beta=0.5, progress=0.8, too_ill_to_move=0.8)
>>> R_progress = 0.5
>>> lurgy.add("R", progress=R_progress)
>>> for i in range(1, 11):
... lurgy.add(f"R{i}", beta=0.0, progress=R_progress)
>>> lurgy.to_json("lurgy6.json", indent=2, auto_bzip=False)
or, in R/RStudio you could type;
> library(metawards)
> lurgy <- metawards$Disease("lurgy6")
> lurgy$add("E", beta=0.0, progress=1.0)
> lurgy$add("I1", beta=0.4, progress=0.2)
> lurgy$add("I2", beta=0.5, progress=0.5, too_ill_to_move=0.5)
> lurgy$add("I3", beta=0.5, progress=0.8, too_ill_to_move=0.8)
> R_progress <- 0.5
> lurgy$add("R", progress=R_progress)
> for(i in 1:10) {
stage <- sprintf("R%d", i)
lurgy$add(stage, beta=0.0, progress=R_progress)
}
> lurgy$to_json("lurgy6.json", indent=2, auto_bzip=False)
or simply copy the below into lurgy6.json;
{
"name": "lurgy6",
"stage": ["E", "I1", "I2", "I3", "R", "R1", "R2",
"R3", "R4", "R5", "R6", "R7", "R8",
"R9", "R10"],
"mapping": ["E", "I", "I", "I", "R", "R", "R",
"R", "R", "R", "R", "R", "R", "R",
"R"],
"beta": [0.0, 0.4, 0.5, 0.5, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
"progress": [1.0, 0.2, 0.5, 0.8, 0.5, 0.5, 0.5,
0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5,
0.5],
"too_ill_to_move": [0.0, 0.0, 0.5, 0.8, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0],
"contrib_foi": [1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0,
1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0],
"start_symptom": 2,
"is_infected": [true, true, true, true, false, false,
false, false, false, false, false, false,
false, false, false]
}
This creates 10 post-recovery stages, with a progress parameter for these stages of 0.5. This means, at quickest, an individual would take 10 days to progress from R to R10, but on average, this will take much longer (as 50% move from one stage to the next each day).
Moving from R to S
Next, we need to add a move function that will move a fraction of individuals from R10 back to S, to represent that fraction losing immunity.
Create a mover called move_immunity.py and copy in the below;
from metawards.movers import MoveGenerator, go_ward, MoveRecord
from metawards.utils import Console
def move_immunity(**kwargs):
def go_immunity(**kwargs):
record = MoveRecord()
gen = MoveGenerator(from_stage="R10", to_stage="S",
fraction=0.5)
go_ward(generator=gen, record=record, **kwargs)
if len(record) > 0:
nlost = record[0][-1]
Console.print(f"{nlost} individual(s) lost immunity today")
return [go_immunity]
This will move 50% of R10 individuals back to S each day.
Note
We have used MoveRecord to record the moves
performed by go_ward(). This keeps a complete
record of exactly how many individuals were moved, and the full
details of that move. In this case, there will only be a single
move (record[0]), and the number of individuals who were moved
is the last value in the record (record[0][-1]).
You can run the model using;
metawards -d lurgy6.json -m single -a 5 --move move_immunity.py
(using the single-ward model, seeding with 5 initial infection).
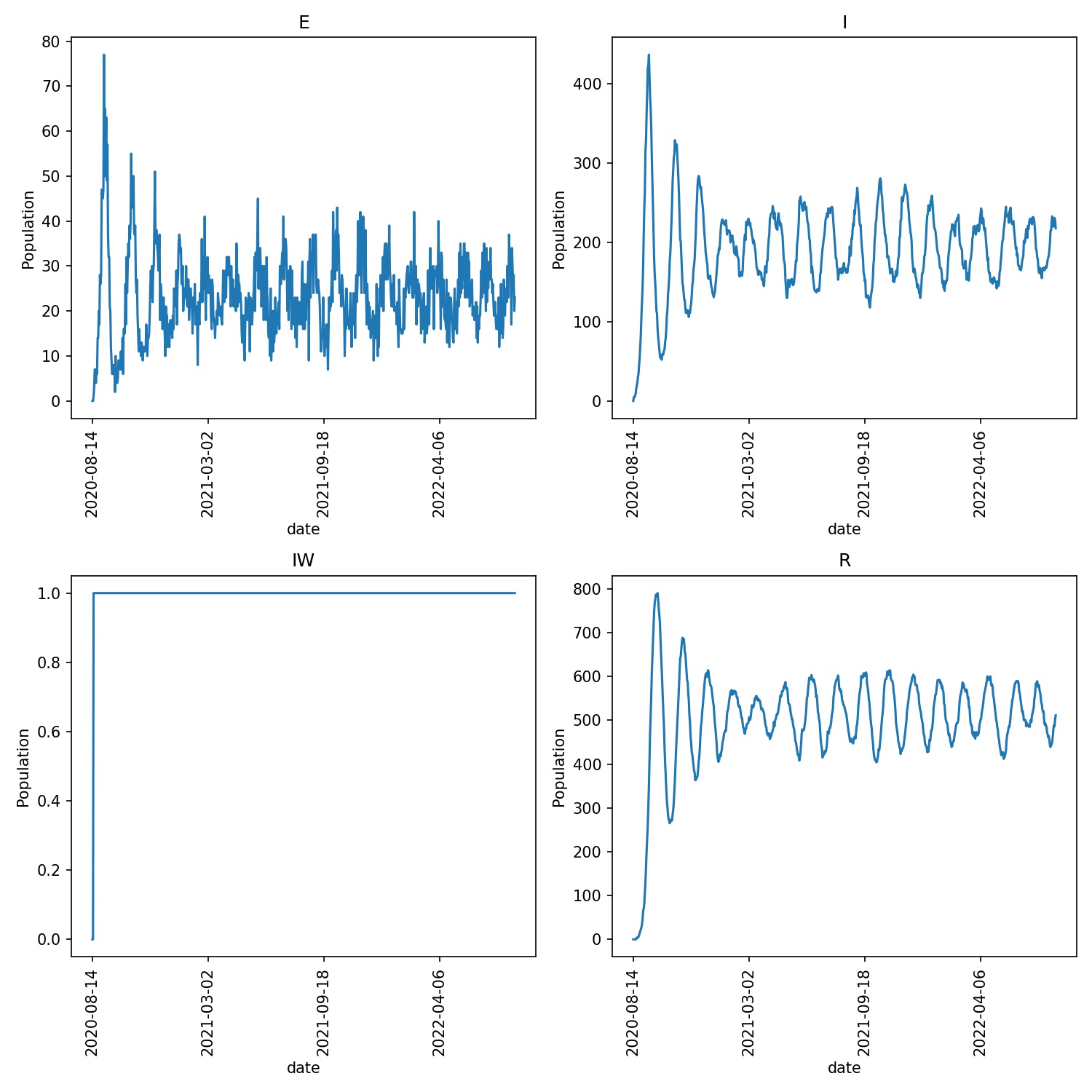
You should see that the outbreak oscillates as individuals who have
lost immunity are re-infected. For example, the graph I get
(from metawards-plot) are shown below;
Note
This is just an illustrative example. Individuals lose immunity in
this model far more quickly than would be expected for a real disease.
You could modify this example to use
custom user variables to
scan through different values of progress for each
of the post-recovery stages, to better model a more realistic disease.
Vaccination and boosters
You can apply the same method to model fading immunity after a vaccination. This could be used to best plan how often booster doses should be deployed.
To do this, we will modify our lurgy to model to include vaccination and post-vaccination stages. For example, in Python (in ipython/Jupyter);
>>> from metawards import Disease
>>> lurgy = Disease("lurgy7")
>>> lurgy.add("E", beta=0.0, progress=1.0)
>>> lurgy.add("I1", beta=0.4, progress=0.2)
>>> lurgy.add("I2", beta=0.5, progress=0.5, too_ill_to_move=0.5)
>>> lurgy.add("I3", beta=0.5, progress=0.8, too_ill_to_move=0.8)
>>> R_progress = 0.5
>>> V_progress = 0.5
>>> lurgy.add("R", progress=R_progress)
>>> for i in range(1, 10):
... lurgy.add(f"R{i}", beta=0.0, progress=R_progress)
>>> lurgy.add("R10", beta=0.0, progress=0.0)
>>> lurgy.add("V", progress=V_progress, is_infected=False)
>>> for i in range(1, 11):
... lurgy.add(f"V{i}", beta=0.0, progress=V_progress,
... is_infected=False)
>>> lurgy.to_json("lurgy7.json", auto_bzip=False)
> library(metawards)
> lurgy <- metawards$Disease("lurgy7")
> lurgy$add("E", beta=0.0, progress=1.0)
> lurgy$add("I1", beta=0.4, progress=0.2)
> lurgy$add("I2", beta=0.5, progress=0.5, too_ill_to_move=0.5)
> lurgy$add("I3", beta=0.5, progress=0.8, too_ill_to_move=0.8)
> R_progress <- 0.5
> V_progress <- 0.5
> lurgy$add("R", progress=R_progress)
> for(i in 1:9) {
stage <- sprintf("R%d", i)
lurgy$add(stage, beta=0.0, progress=R_progress)
}
> lurgy.add("R10", beta=0.0, progress=0.0)
> lurgy.add("V", progress=V_progress, is_infected=False)
> for(i in 1:10) {
stage <- sprintf("V%d", i)
lurgy$add(stage, beta=0.0, progress=V_progress)
}
> lurgy.to_json("lurgy7.json", auto_bzip=False)
or copy the below into lurgy7.json
{
"name": "lurgy7",
"stage": ["E", "I1", "I2", "I3", "R", "R1",
"R2", "R3", "R4", "R5", "R6", "R7",
"R8", "R9", "R10", "V", "V1", "V2",
"V3", "V4", "V5", "V6", "V7", "V8",
"V9", "V10"],
"mapping": ["E", "I", "I", "I", "R", "R", "R",
"R", "R", "R", "R", "R", "R", "R",
"R", "V", "V", "V", "V", "V", "V",
"V", "V", "V", "V", "V"],
"beta": [0.0, 0.4, 0.5, 0.5, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0],
"progress": [1.0, 0.2, 0.5, 0.8, 0.5, 0.5,
0.5, 0.5, 0.5, 0.5, 0.5, 0.5,
0.5, 0.5, 0.0, 0.5, 0.5, 0.5,
0.5, 0.5, 0.5, 0.5, 0.5, 0.5,
0.5, 0.5],
"too_ill_to_move": [0.0, 0.0, 0.5, 0.8, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0],
"contrib_foi": [1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0,
1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0,
1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0,
1.0, 1.0, 1.0, 1.0, 1.0],
"start_symptom": 2,
"is_infected": [true, true, true, true, false, false,
false, false, false, false, false, false,
false, false, false, false, false, false,
false, false, false, false, false, false,
false, false]
}
Note
Note how progress for the R10 stage is set to 0 to prevent
R10 automatic progression from R10 to V.
Note
Note also that we have to manually set is_infected to false for
the V stages. This is set automatically to false only for R stages.
Next, modify move_immunity.py to read;
from metawards.movers import MoveGenerator, go_ward, MoveRecord
from metawards.utils import Console
def move_immunity(**kwargs):
def go_vaccinate(population, **kwargs):
if population.day <= 10:
gen = MoveGenerator(from_stage="S", to_stage="V",
number=100)
go_ward(generator=gen, population=population, **kwargs)
def go_immunity(**kwargs):
record = MoveRecord()
gen = MoveGenerator(from_stage="R10", to_stage="S",
fraction=0.5)
go_ward(generator=gen, record=record, **kwargs)
if len(record) > 0:
Console.print(f"{record[0][-1]} individual(s) lost immunity today")
def go_booster(**kwargs):
gen = MoveGenerator(from_stage="V10", to_stage="V",
fraction=0.2)
go_ward(generator=gen, **kwargs)
record = MoveRecord()
gen = MoveGenerator(from_stage="V10", to_stage="S")
go_ward(generator=gen, record=record, **kwargs)
if len(record) > 0:
Console.print(f"{record[0][-1]} individual(s) didn't get their booster")
return [go_vaccinate, go_booster, go_immunity]
Here, we’ve added a go_vaccinate function that, for the first
10 days of the outbreak, moves up to 100 individuals
per day from S to V. This will, in effect, vaccinate all of the
population of 1000 individuals who are not infected.
Next, we’ve added a go_booster function that samples 20% of the V10
stage to give them a booster vaccine that returns them to V. The
remaining individuals in V10 miss their booster dose, and are
returned to S.
You can run the model using;
metawards -d lurgy7.json -m single -a 5 --move move_immunity.py
You should see that the infection nearly dies out, as nearly everyone is vaccinated. However, a small number of lingering infections spark a second outbreak amongst individuals who miss their booster shot, leading then to a cycle of infection and losing immunity, e.g.
─────────────────────────────────────────────── Day 9 ────────────────────────────────────────────────
S: 89 E: 0 I: 6 V: 900 R: 5 IW: 0 POPULATION: 1000
Number of infections: 6
─────────────────────────────────────────────── Day 10 ───────────────────────────────────────────────
S: 0 E: 0 I: 6 V: 989 R: 5 IW: 0 POPULATION: 1000
Number of infections: 6
─────────────────────────────────────────────── Day 11 ───────────────────────────────────────────────
S: 0 E: 0 I: 6 V: 989 R: 5 IW: 0 POPULATION: 1000
Number of infections: 6
─────────────────────────────────────────────── Day 12 ───────────────────────────────────────────────
1 individual(s) didn't get their booster
S: 1 E: 0 I: 6 V: 988 R: 5 IW: 0 POPULATION: 1000
Number of infections: 6
...
─────────────────────────────────────────────── Day 23 ───────────────────────────────────────────────
54 individual(s) didn't get their booster
S: 309 E: 0 I: 2 V: 680 R: 9 IW: 0 POPULATION: 1000
Number of infections: 2
─────────────────────────────────────────────── Day 24 ───────────────────────────────────────────────
72 individual(s) didn't get their booster
2 individual(s) lost immunity today
S: 383 E: 0 I: 2 V: 608 R: 7 IW: 0 POPULATION: 1000
Number of infections: 2
─────────────────────────────────────────────── Day 25 ───────────────────────────────────────────────
60 individual(s) didn't get their booster
S: 443 E: 0 I: 2 V: 548 R: 7 IW: 0 POPULATION: 1000
Number of infections: 2
─────────────────────────────────────────────── Day 26 ───────────────────────────────────────────────
64 individual(s) didn't get their booster
1 individual(s) lost immunity today
S: 507 E: 1 I: 2 V: 484 R: 6 IW: 1 POPULATION: 1000
Number of infections: 3
─────────────────────────────────────────────── Day 27 ───────────────────────────────────────────────
49 individual(s) didn't get their booster
1 individual(s) lost immunity today
S: 557 E: 0 I: 3 V: 435 R: 5 IW: 0 POPULATION: 1000
Number of infections: 3
─────────────────────────────────────────────── Day 28 ───────────────────────────────────────────────
44 individual(s) didn't get their booster
S: 599 E: 2 I: 3 V: 391 R: 5 IW: 1 POPULATION: 1000
Number of infections: 5
...
─────────────────────────────────────────────── Day 40 ───────────────────────────────────────────────
8 individual(s) didn't get their booster
S: 776 E: 7 I: 39 V: 162 R: 16 IW: 1 POPULATION: 1000
Number of infections: 46
─────────────────────────────────────────────── Day 41 ───────────────────────────────────────────────
8 individual(s) didn't get their booster
S: 776 E: 8 I: 45 V: 154 R: 17 IW: 1 POPULATION: 1000
Number of infections: 53
...
────────────────────────────────────────────── Day 103 ───────────────────────────────────────────────
11 individual(s) lost immunity today
S: 289 E: 32 I: 329 V: 2 R: 348 IW: 1 POPULATION: 1000
Number of infections: 361
────────────────────────────────────────────── Day 104 ───────────────────────────────────────────────
12 individual(s) lost immunity today
S: 258 E: 43 I: 319 V: 2 R: 378 IW: 1 POPULATION: 1000
Number of infections: 362
────────────────────────────────────────────── Day 105 ───────────────────────────────────────────────
1 individual(s) didn't get their booster
11 individual(s) lost immunity today
S: 233 E: 37 I: 325 V: 1 R: 404 IW: 1 POPULATION: 1000
Number of infections: 362
Note
Again, this is just an illustrative example. Immunity from vaccination would be expected to last for much longer than a couple of weeks. You could use adjustable variables (and custom user-adjustable variables) to scan through the progress along the post-vaccination stages, the numbers vaccinated each day, and different percentages of individuals who take a booster, to better model a real situation.
Note
We have used Disease.progress
to slow movement along the post-recovery and post-vaccinated stages.
An alternative method would be to write a custom iterator to
replace advance_recovery(). This could
slow down movement programmatically, e.g. by only testing for
advancement along the R and V stages every 10 days, as
opposed to every day. We’ve designed metawards to be very
flexible, so that you have many choices for how you want to
model different scenarios.